File IO¶
Examples¶
Download Sample Files¶
Read CSV Example
Read/Write CSV Example

Read NetCDF Example
Read multiple NetCDF Example
Read/Write NetCDF Example
CSV file¶
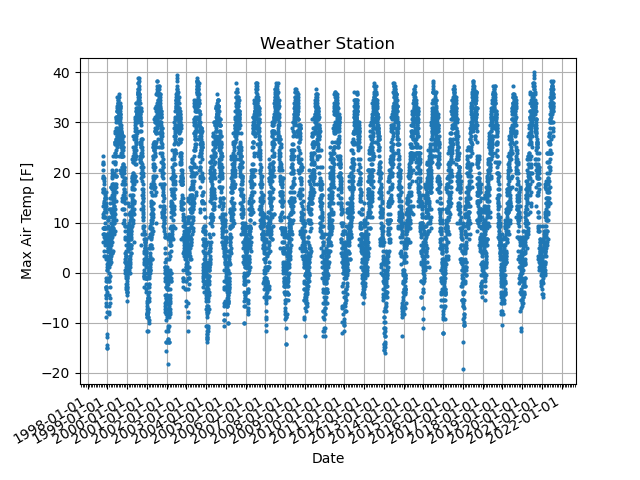
The CSV file is the typical format for weather station data. It stands for the “comma separated value” written by the text (or ASCII) format. It generally consists of data description (called a header) and data value. The package Pandas provides an easy way to read CSV files.
An example of CSV file¶
Let’s try to read the CSV file GHCN_sample.csv.
This CSV file includes a note on Lines 1-15 and a header on Line 16.
After Line 17, we can see the data separated by a comma. The value “M” indicates a missing value.
The data contains day, precipitation, multi-day precipitation, snow depth, snowfall, minimum temperature, maximum temperature, and reference evapotranspiration.
1This data has been provided by the Utah Climate Center. Please cite the Climate Center when using this data.
2Station Network,GHCN:AWOS
3Station ID,USW00094128
4Station Name,LOGAN CACHE AP
5Latitude,41.7872
6Longitude,-111.8533
7Elevation,1357.6 m
8State,UT
9Country,US
10Units,Metric
11*Note: 'S' Indicates a row that has been filled in by the system as a missing day.
12*Note: All data is displayed the same as we receive it. If there are any values that may seem wrong
13then download the data again and select the 'Show source/quality' option to view the flags.
14
15
16Day,Precipitation,Multi-Day Precipitation,Snow Depth,Snow Fall,Min Temperature,Max Temperature,Ref Evapotranspiration
171998-10-01,M,M,M,M,M,M,M
181998-10-02,M,M,M,M,M,M,M
191998-10-03,M,M,M,M,M,M,M
201998-10-04,4.3,M,M,M,0.6,8.9,1.48056
An example of Python script¶
Let’s check the sample file, rw_csv-v2.py, to read and write a CSV file.
General packages¶
1import numpy as np
2import pandas as pd
3import xarray as xr
4import os
5import matplotlib.pyplot as plt
6import matplotlib.dates as mdates
7from datetime import datetime
This script will read the CSV file using Pandas and then get the data as Xarray (and Numpy). There are some date formats in weather station data. For example, we describe August 24, 2021, as of 10-21-2021 or 10/21/21. The “DateTime” package is convenient to adjust such a date format. The “OS” package is also helpful to obtain the filename. This script also displays a plot using the package “Matplotlib,” but it is optional here.
Read a CSV file¶
10# ----- Parameter setting ------
11# == Date ==
12dateparse = lambda x: datetime.strptime(x, '%Y-%m-%d')
13#dateparse = lambda x: datetime.strptime(x, '%Y-%m-%d %H:%M:%S')
14#dateparse = lambda x: datetime.strptime(x, '%m/%d/%y %H:%M')
15
16# == CSV file name and location"
17fcsv = 'GHCN_sample.csv'
18
19# == Your definition of variable names
20## Day,Precipitation, Multi-Day Precipitation, Snow Depth, Snow Fall, Min Temperature, Max Temperature, Ref Evapotranspiration,
21vnames = ['date','precip','mdpr','sdep','sfall','tmin','tmax','refe']
22
23# == location of header. It starts from 0
24nhead = 13
25
26# == Figure name ==
27fnFIG = os.path.splitext(os.path.basename(__file__))[0]
28
29# ------------------------------
30
31print(' ----- Read CSV file ------')
32# additional parameters: encoding = 'ISO-8859-1'
33fin = pd.read_csv(fcsv, \
34 delimiter = ',', header = nhead, na_values = ['M','T','S'],\
35 names = vnames, parse_dates = ['date'], date_parser = dateparse)
36
37
38print(fin)
39print(' ----------------------------')
40
41print(' ----- Time setting ------')
42date = fin.date.astype('datetime64[D]')
43print(date)
44print(' ----------------------------')
45
46print(' ----- Get Maximum Temperature ------')
47tmax = xr.DataArray(fin.tmax.astype(float), dims=('date'), coords={'date': date})
48tmin = xr.DataArray(fin.tmin.astype(float), dims=('date'), coords={'date': date})
This script uses “pandas.read_csv” function to read the CSV file (Line 33-35). There are some options in this function. The option delimiter should be “,” for the CSV file. This script defines the file name (Line 17), the number of lines for the header (Line 24), and the missing value (na_values). It also defines the variable names on Line 21. The variable name in the first column is “date” here. The CSV file shows the date formate of October 1, 1998, as “1998-10-01”. Here, we define the date formate as “%Y-%m-%d” on Line 12, which puts into the option of “parse_dates” and “date_parser” for the function.
Note
Here are examples of date format.
%Y-%m-%d (e.g., 2021-10-13)
%m/%d/%y (e.g., 10/13/21)
%Y-%d-%m %H:%M:%S (e.g., 2021-10-13 00:00:00)
Write a CSV file¶
54df = pd.DataFrame ({'date': date, 'tavg': tavg, 'tmin': tmin, 'tmax': tmax})
55df.to_csv(fnFIG+".csv",index=False)
56print(df)
To write a CSV file, we use a Pandas function pandas.DataFrame.to_csv. An output file name has the same file name of this script as defined on Line 27, but for the CSV file extension (“.csv”). The output CSV file looks like below.
1date,tavg,tmin,tmax
21998-10-01,,,
31998-10-02,,,
41998-10-03,,,
51998-10-04,4.75,0.6,8.9
NetCDF file¶
Read and write a single NetCDF file¶
Let’s check the sample file, rw_netcdf-v2.py, to read and write a NetCDF file.
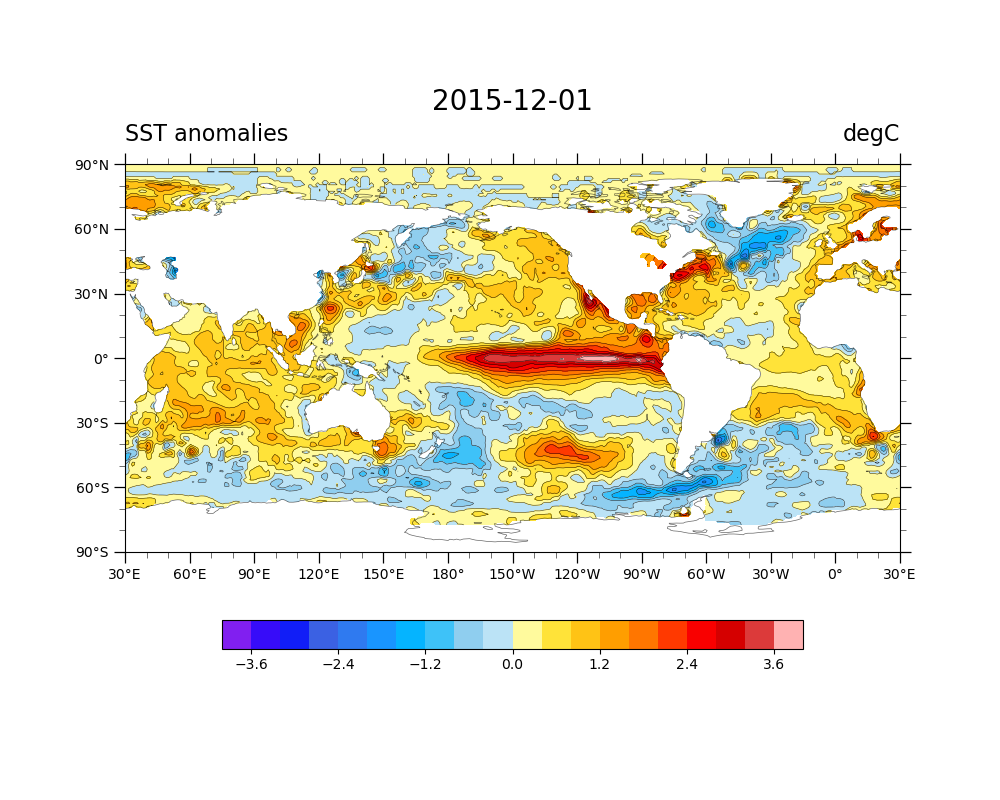
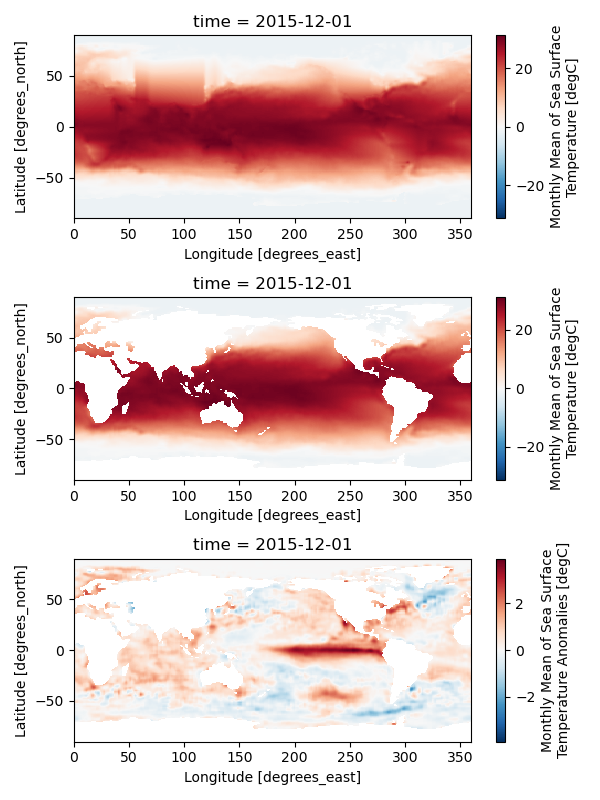
This script reads monthly sea surface temperature provided by NOAA (oisst_monthly.nc).
It also requires the land-sea mask file (lsmask.nc).
General packages¶
1import numpy as np
2import pandas as pd
3import xarray as xr
4import matplotlib.pyplot as plt
5import matplotlib.dates as mdates
6from datetime import datetime
This script uses Xarray to read and write the NetCDF file. As a default setting, Xarray may not support the NetCDF. In that case, you need to install other packages, such as Dask, NetCDF4, or PyNIO. Please check Install Packages how to install NetCDF.
Read a NetCDF file¶
8# ----- Parameter setting ------
9
10# == Figure name ==
11fnFIG = 'rw_netcdf-v2.png'
12
13sdate = '1982-01-01'
14edate = '2020-12-01'
15
16# -- Draw date
17ddate = '2015-12-01'
18
19# == netcdf file name and location"
20fnc = 'oisst_monthly.nc'
21dmask = xr.open_dataset('lsmask.nc')
22print(dmask)
23
24
25# == read netcdf data
26ds = xr.open_dataset(fnc)
27print(ds)
28
29sst = ds.sst.where(dmask.mask.isel(time=0) == 1)
30
31clm = sst.sel(time=slice(sdate,edate)).groupby('time.month').mean(dim='time')
32anm = (sst.groupby('time.month') - clm)
33#print(clm)
34#print(anm)
35
To read a signle NetCDF file, we use a xarray function xarray.open_dataset, as described on Lines 21 and 26. The resultant varialbe “ds” is the DataSet array. You can extract the sea surface temperature DataArray from “ds” on Line 29. We also applied the land-sea mask to obain the sea surface temperature over the ocean not the land using a DataArray.where function.
Read multiple NetCDF files¶
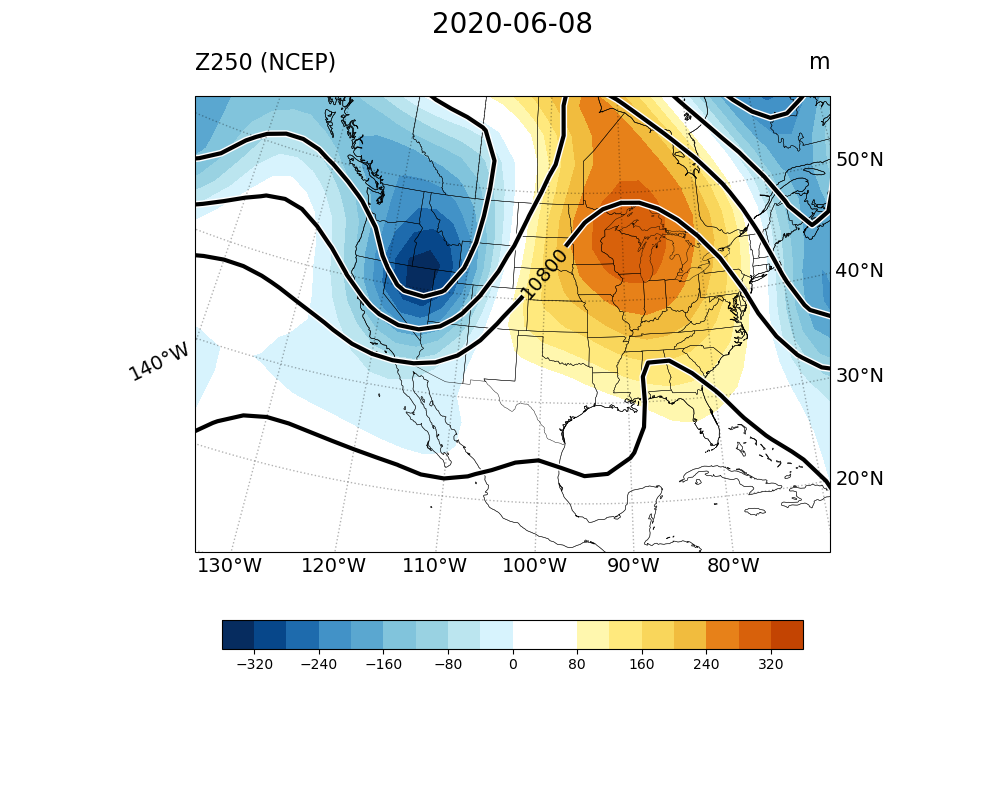
To read multiple NetCDF file, we use the differnet xarray function xarray.open_mfdataset. Following is an example to read the mulple NetCDF file and extract geopotential height at 250 hPa as the DataArray “dat”.
Sample file read-multi_netcdf-v2.py
23# == netcdf file name and location"
24fnc = "hgt_ncep_daily.*.nc"
25
26ds = xr.open_mfdataset(fnc, parallel=True)
27print(ds)
28
29dat = ds.hgt.sel(level=250)
30print(dat)