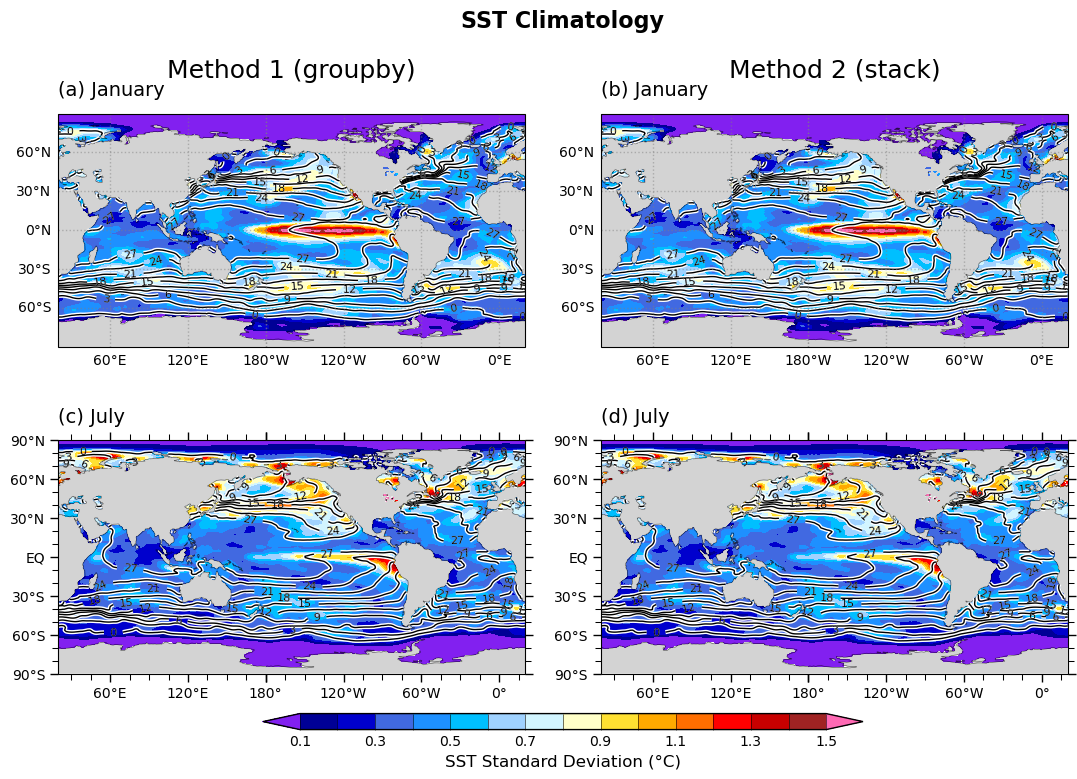
Global Map (Climatology)#
# --- Packages ---
## General Packages
import pandas as pd
import xarray as xr
import numpy as np
import os
import ipynbname
## GeoCAT
import geocat.comp as gccomp
import geocat.viz as gv
import geocat.viz.util as gvutil
## Visualization
import cmaps
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import shapely.geometry as sgeom
## MatPlotLib
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
import matplotlib.patches as mpatches
import matplotlib.dates as mdates
from matplotlib.colors import ListedColormap, BoundaryNorm
from matplotlib.ticker import MultipleLocator
# --- Automated output filename ---
def get_filename():
try:
# Will fail when __file__ is undefined (e.g., in notebooks)
filename = os.path.splitext(os.path.basename(__file__))[0]
except NameError:
try:
# Will fail during non-interactive builds
import ipynbname
nb_path = ipynbname.path()
filename = os.path.splitext(os.path.basename(str(nb_path)))[0]
except Exception:
# Fallback during Jupyter Book builds or other headless execution
filename = "template_analysis_clim"
return filename
fnFIG = get_filename() + ".png"
print(f"Figure filename: {fnFIG}")
Figure filename: template_analysis_clim.png
# --- Parameter setting ---
data_dir="../data"
fname = "sst.cobe2.185001-202504.nc"
ystr, yend = 1991, 2020
fvar = "sst"
# --- Reading NetCDF Dataset ---
# Construct full path and open dataset
path_data = os.path.join(data_dir, fname)
ds = xr.open_dataset(path_data)
# Extract the variable
var = ds[fvar]
# Ensure dimensions are (time, lat, lon)
var = var.transpose("time", "lat", "lon", missing_dims="ignore")
# Ensure latitude is ascending
if var.lat.values[0] > var.lat.values[-1]:
var = var.sortby("lat")
# Ensure time is in datetime64 format
if not np.issubdtype(var.time.dtype, np.datetime64):
try:
var["time"] = xr.decode_cf(ds).time
except Exception as e:
raise ValueError("Time conversion to datetime64 failed: " + str(e))
# === Select time range ===
dat = var.sel(time=slice(f"{ystr}-01-01", f"{yend}-12-31"))
print(dat)
<xarray.DataArray 'sst' (time: 360, lat: 180, lon: 360)> Size: 93MB
[23328000 values with dtype=float32]
Coordinates:
* lat (lat) float32 720B -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float32 1kB 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
* time (time) datetime64[ns] 3kB 1991-01-01 1991-02-01 ... 2020-12-01
Attributes:
long_name: Monthly Means of Global Sea Surface Temperature
valid_range: [-5. 40.]
units: degC
var_desc: Sea Surface Temperature
dataset: COBE-SST2 Sea Surface Temperature
statistic: Mean
parent_stat: Individual obs
level_desc: Surface
actual_range: [-3.0000002 34.392 ]
# --- Climatology, Anomalies, and Standard Deviation
# Using groupby
clm1 = dat.groupby("time.month").mean(dim="time")
anm1 = dat.groupby("time.month") - clm1
std1 = anm1.groupby("time.month").std(dim="time")
print(clm1.sizes)
print(anm1.sizes)
print(std1.sizes)
Frozen({'month': 12, 'lat': 180, 'lon': 360})
Frozen({'time': 360, 'lat': 180, 'lon': 360})
Frozen({'month': 12, 'lat': 180, 'lon': 360})
# Using stack and unstack to convert from time to year x month
dat = dat.assign_coords(year=dat.time.dt.year, month=dat.time.dt.month)
dat4D = dat.set_index(time=["year", "month"]).unstack("time")
dat4D = dat4D.transpose("year", "month", "lat", "lon")
print(dat4D.sizes)
# Calculate climatology, anomaly, and standar deviations
clm2 = dat4D.mean(dim="year")
anm2 = dat4D-clm2
std2 = anm2.std(dim="year")
print(clm2.sizes)
print(anm2.sizes)
print(std2.sizes)
Frozen({'year': 30, 'month': 12, 'lat': 180, 'lon': 360})
Frozen({'month': 12, 'lat': 180, 'lon': 360})
Frozen({'year': 30, 'month': 12, 'lat': 180, 'lon': 360})
Frozen({'month': 12, 'lat': 180, 'lon': 360})
def plot_global_panel_OP1(ax, std_data, std_levels, titleM, titleL, color_list,
clm_data=None, clm_levels=None):
"""
Plot global map of monthly standard deviation with optional climatology contours.
Parameters
----------
ax : cartopy.mpl.geoaxes.GeoAxesSubplot
Axes to plot on.
std_data : xarray.DataArray (month, lat, lon)
Standard deviation data to plot with filled contours.
std_levels : list or array-like
Contour levels for the std_data.
title : str
Title above the plot panel.
color_list : list
Colors for the colormap.
clm_data : xarray.DataArray (month, lat, lon), optional
Climatology data for contour overlay.
clm_levels : list or array-like, optional
Contour levels for climatology.
"""
# --- Add cyclic longitudes ---
std_cyclic = gvutil.xr_add_cyclic_longitudes(std_data, "lon")
# --- Plot standard deviation ---
im = ax.contourf(
std_cyclic["lon"],
std_cyclic["lat"],
std_cyclic,
levels=std_levels,
cmap=color_list,
extend="both",
transform=ccrs.PlateCarree()
)
# --- Plot climatology contours (optional) ---
if clm_data is not None:
clm_cyclic = gvutil.xr_add_cyclic_longitudes(clm_data, "lon")
levels = clm_levels if clm_levels is not None else 10
csW = ax.contour(
clm_cyclic["lon"], clm_cyclic["lat"], clm_cyclic,
levels=levels, colors="white", linewidths=3,
transform=ccrs.PlateCarree()
)
csB = ax.contour(
clm_cyclic["lon"], clm_cyclic["lat"], clm_cyclic,
levels=levels, colors="black", linewidths=1,
transform=ccrs.PlateCarree()
)
ax.clabel(csW, inline=1, fontsize=8, fmt="%.0f")
ax.clabel(csB, inline=1, fontsize=8, fmt="%.0f")
# --- Map features ---
ax.add_feature(cfeature.LAND, facecolor='lightgray', edgecolor='none', zorder=0)
ax.coastlines(linewidth=0.5, alpha=0.6)
ax.set_extent([-180, 180, -90, 90], crs=ccrs.PlateCarree())
# --- Option 1 ---
gl = ax.gridlines(draw_labels=True, linestyle='dotted', color='gray', linewidth=1, alpha=0.5)
gl.top_labels = False
gl.right_labels = False
gl.xlines = True
gl.ylines = True
# Set major tick locations
gl.xlocator = mticker.FixedLocator(np.arange(-180, 181, 60))
gl.ylocator = mticker.FixedLocator(np.arange(-90, 91, 30))
# Optional: Format labels with degree symbols (default is just numbers)
gl.xlabel_style = {"size": 10}
gl.ylabel_style = {"size": 10}
gl.xformatter = mticker.FuncFormatter(lambda x, _: f"{abs(int(x))}°{'E' if x >= 0 else 'W'}")
gl.yformatter = mticker.FuncFormatter(lambda y, _: f"{abs(int(y))}°{'N' if y >= 0 else 'S'}")
gvutil.set_titles_and_labels(ax, maintitle=titleM, maintitlefontsize=16, lefttitle=titleL, lefttitlefontsize=14)
return im
def plot_global_panel_OP2(ax, std_data, std_levels, titleM, titleL, color_list,
clm_data=None, clm_levels=None):
"""
Plot global map of monthly standard deviation with optional climatology contours.
Parameters
----------
ax : cartopy.mpl.geoaxes.GeoAxesSubplot
Axes to plot on.
std_data : xarray.DataArray (month, lat, lon)
Standard deviation data to plot with filled contours.
std_levels : list or array-like
Contour levels for the std_data.
title : str
Title above the plot panel.
color_list : list
Colors for the colormap.
clm_data : xarray.DataArray (month, lat, lon), optional
Climatology data for contour overlay.
clm_levels : list or array-like, optional
Contour levels for climatology.
"""
# --- Add cyclic longitudes ---
std_cyclic = gvutil.xr_add_cyclic_longitudes(std_data, "lon")
# --- Plot standard deviation ---
im = ax.contourf(
std_cyclic["lon"],
std_cyclic["lat"],
std_cyclic,
levels=std_levels,
cmap=color_list,
extend="both",
transform=ccrs.PlateCarree()
)
# --- Plot climatology contours (optional) ---
if clm_data is not None:
clm_cyclic = gvutil.xr_add_cyclic_longitudes(clm_data, "lon")
levels = clm_levels if clm_levels is not None else 10
csW = ax.contour(
clm_cyclic["lon"], clm_cyclic["lat"], clm_cyclic,
levels=levels, colors="white", linewidths=3,
transform=ccrs.PlateCarree()
)
csB = ax.contour(
clm_cyclic["lon"], clm_cyclic["lat"], clm_cyclic,
levels=levels, colors="black", linewidths=1,
transform=ccrs.PlateCarree()
)
ax.clabel(csW, inline=1, fontsize=8, fmt="%.0f")
ax.clabel(csB, inline=1, fontsize=8, fmt="%.0f")
# --- Map features ---
ax.add_feature(cfeature.LAND, facecolor='lightgray', edgecolor='none', zorder=0)
ax.coastlines(linewidth=0.5, alpha=0.6)
ax.set_extent([-180, 180, -90, 90], crs=ccrs.PlateCarree())
# --- Option 2: Manual setting ---
# --- Set major ticks explicitly ---
ax.set_xticks(np.arange(-180, 181, 60), crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(-90, 91, 30), crs=ccrs.PlateCarree())
# Custom tick labels
ax.set_yticklabels(["90°S", "60°S", "30°S", "EQ", "30°N", "60°N", "90°N"])
ax.set_xticklabels(["180°", "120°W", "60°W", "0°", "60°E", "120°E", ""])
# --- Set minor tick locators ---
ax.xaxis.set_minor_locator(mticker.MultipleLocator(15))
ax.yaxis.set_minor_locator(mticker.MultipleLocator(10))
# --- Add tick marks ---
gvutil.add_major_minor_ticks(
ax,
labelsize=10,
x_minor_per_major=4, # 15° spacing for minor if major is 60°
y_minor_per_major=3 # 10° spacing for minor if major is 30°
)
# --- Customize ticks to show outward ---
ax.tick_params(
axis="both",
which="both",
direction="out",
length=6,
width=1,
top=True,
bottom=True,
left=True,
right=True
)
ax.tick_params(
axis="both",
which="minor",
length=4,
width=0.75
)
gvutil.set_titles_and_labels(ax, maintitle=titleM, maintitlefontsize=16, lefttitle=titleL, lefttitlefontsize=14)
return im
# --- FIGURE PLOT ---
# === Global settings ===
proj = ccrs.PlateCarree(central_longitude=200)
fig, axes = plt.subplots(nrows=2, ncols=2, figsize=(12, 8), subplot_kw={'projection': proj})
fig.suptitle("SST Climatology", fontsize=16, fontweight="bold")
axes = axes.flatten() # 🔑 Make it a 1D list of axes
colormap = cmaps.amwg_blueyellowred.colors # Use the AMWG BlueYellowRed colormap
color_list = ListedColormap(colormap[1:15]) # Use 14 colors (excluding the most extreme ends)
# Set distinct colors for under and over
color_list.set_under(colormap[0]) # Use the first color for underflow (< first level)
color_list.set_over(colormap[15]) # Use the last color for overflow (> last level)
clevs_std = np.linspace(0.1, 1.5, 15) # 15 intervals → 16 colors
clevs_clm = np.arange(0, 36, 3)
# === Plot panels ===
imon = 1 # January
ip=0
im=plot_global_panel_OP1(axes[ip],
std_data=std1.sel(month=imon),
std_levels=clevs_std,
titleM="Method 1 (groupby)",
titleL="(a) January",
color_list=color_list,
clm_data=clm1.sel(month=imon),
clm_levels=clevs_clm
)
ip=1
plot_global_panel_OP1(axes[ip],
std_data=std2.sel(month=imon),
std_levels=clevs_std,
titleM="Method 2 (stack)",
titleL="(b) January",
color_list=color_list,
clm_data=clm2.sel(month=imon),
clm_levels=clevs_clm
)
imon = 7 # July
ip=2
plot_global_panel_OP2(axes[ip],
std_data=std1.sel(month=imon),
std_levels=clevs_std,
titleM="",
titleL="(c) July",
color_list=color_list,
clm_data=clm1.sel(month=imon),
clm_levels=clevs_clm
)
ip=3
plot_global_panel_OP2(axes[ip],
std_data=std2.sel(month=imon),
std_levels=clevs_std,
titleM="",
titleL="(d) July",
color_list=color_list,
clm_data=clm2.sel(month=imon),
clm_levels=clevs_clm
)
# --- Colorbar for im1 ---
cbar_ax = fig.add_axes([0.25, 0.08, 0.5, 0.02])
cbar = plt.colorbar(im, cax=cbar_ax, orientation="horizontal",
spacing="uniform", extend='both',
extendfrac=0.07, # 🔑 Make the edge triangles slightly larger
drawedges=True)
cbar.set_label("SST Standard Deviation (°C)", fontsize=12)
cbar.ax.tick_params(length=0, labelsize=10)
cbar.outline.set_edgecolor("black")
cbar.outline.set_linewidth(1.0)
# === Finalize ===
#plt.tight_layout()
plt.subplots_adjust(hspace=0.4, wspace=0.01, left=0.05, right=0.95, top=0.85, bottom=0.15)
plt.savefig(fnFIG, dpi=300, bbox_inches="tight")
plt.show()