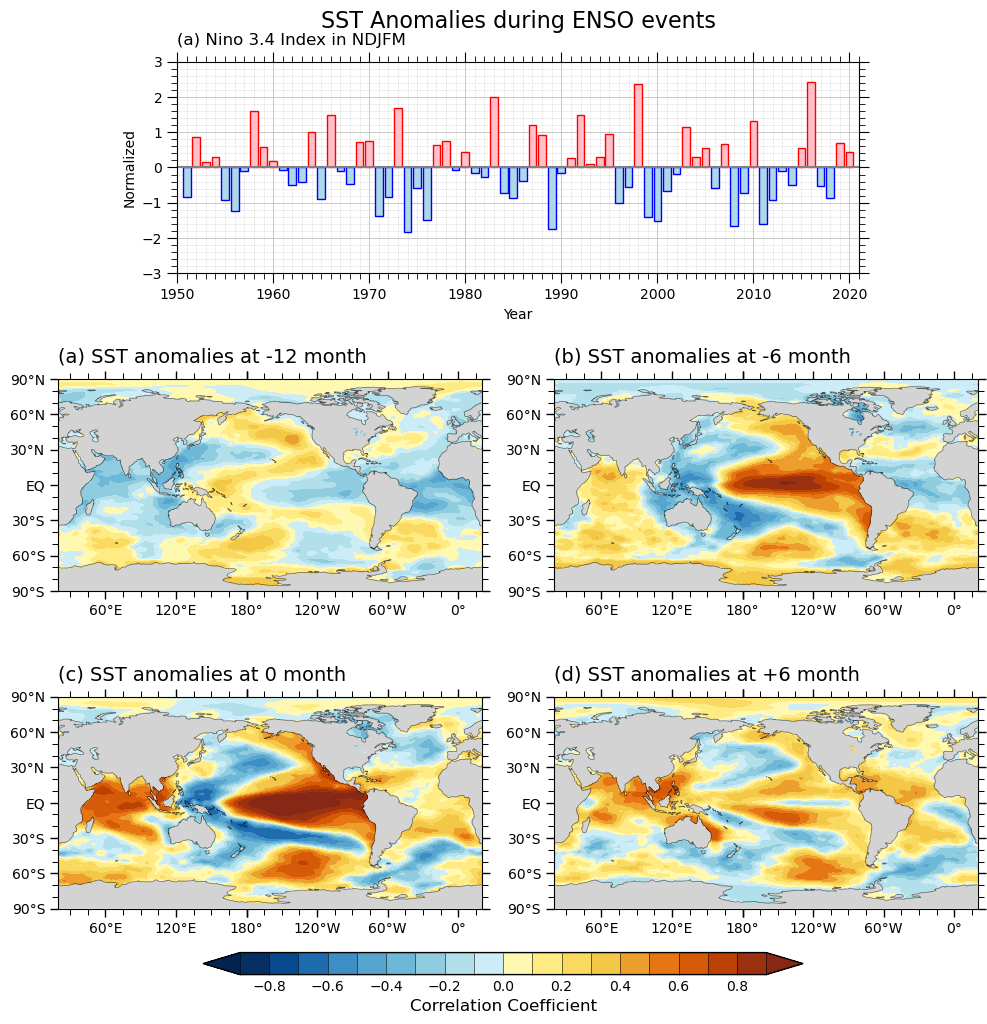
Global Map (ENSO)#
# --- Packages ---
## General Packages
import pandas as pd
import xarray as xr
import numpy as np
import os
import ipynbname
## GeoCAT
import geocat.comp as gccomp
import geocat.viz as gv
import geocat.viz.util as gvutil
## Visualization
import cmaps
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import shapely.geometry as sgeom
## MatPlotLib
import matplotlib.pyplot as plt
import matplotlib.ticker as mticker
import matplotlib.patches as mpatches
import matplotlib.dates as mdates
from matplotlib.colors import ListedColormap, BoundaryNorm
from matplotlib.ticker import MultipleLocator
## Unique Plots
import matplotlib.gridspec as gridspec
# --- Automated output filename ---
def get_filename():
try:
# Will fail when __file__ is undefined (e.g., in notebooks)
filename = os.path.splitext(os.path.basename(__file__))[0]
except NameError:
try:
# Will fail during non-interactive builds
import ipynbname
nb_path = ipynbname.path()
filename = os.path.splitext(os.path.basename(str(nb_path)))[0]
except Exception:
# Fallback during Jupyter Book builds or other headless execution
filename = "template_analysis_clim"
return filename
fnFIG = get_filename() + ".png"
print(f"Figure filename: {fnFIG}")
Figure filename: template_analysis_clim.png
# --- Parameter setting ---
data_dir="../data"
fname = "sst.cobe2.185001-202504.nc"
ystr, yend = 1950, 2020
fvar = "sst"
# --- Reading NetCDF Dataset ---
# Construct full path and open dataset
path_data = os.path.join(data_dir, fname)
ds = xr.open_dataset(path_data)
# Extract the variable
var = ds[fvar]
# Ensure dimensions are (time, lat, lon)
var = var.transpose("time", "lat", "lon", missing_dims="ignore")
# Ensure latitude is ascending
if var.lat.values[0] > var.lat.values[-1]:
var = var.sortby("lat")
# Ensure time is in datetime64 format
if not np.issubdtype(var.time.dtype, np.datetime64):
try:
var["time"] = xr.decode_cf(ds).time
except Exception as e:
raise ValueError("Time conversion to datetime64 failed: " + str(e))
# === Select time range ===
dat = var.sel(time=slice(f"{ystr}-01-01", f"{yend}-12-31"))
print(dat)
<xarray.DataArray 'sst' (time: 852, lat: 180, lon: 360)> Size: 221MB
[55209600 values with dtype=float32]
Coordinates:
* lat (lat) float32 720B -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float32 1kB 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
* time (time) datetime64[ns] 7kB 1950-01-01 1950-02-01 ... 2020-12-01
Attributes:
long_name: Monthly Means of Global Sea Surface Temperature
valid_range: [-5. 40.]
units: degC
var_desc: Sea Surface Temperature
dataset: COBE-SST2 Sea Surface Temperature
statistic: Mean
parent_stat: Individual obs
level_desc: Surface
actual_range: [-3.0000002 34.392 ]
def calc_seasonal_mean(dat, window=5, end_month=1, min_coverage=0.9, dtrend=True):
"""
Calculate seasonal mean using a trailing running mean, extract the final month (e.g., Mar for NDJFM),
convert to year-lat-lon DataArray, apply minimum coverage mask, and optionally remove trend.
Parameters:
-----------
dat : xr.DataArray
Input data with dimensions (time, lat, lon) and datetime64 'time'.
window : int
Running mean window size (default is 5).
end_month : int
Target month used to extract seasonal means (final month of the trailing average).
min_coverage : float
Minimum fraction of year coverage required for masking (default 0.9).
dtrend : bool
If True, remove linear trend after applying coverage mask.
Returns:
--------
dat_out : xr.DataArray
Seasonal mean with dimensions (year, lat, lon), optionally detrended and masked.
"""
# Compute monthly anomalies
clm = dat.groupby("time.month").mean(dim="time")
anm = dat.groupby("time.month") - clm
# Apply trailing running mean
dat_rm = anm.rolling(time=window, center=False, min_periods=window).mean()
# Filter for entries where month == end_month
dat_tmp = dat_rm.sel(time=dat_rm["time"].dt.month == end_month)
# Extract year from the end_month timestamps
years = dat_tmp["time"].dt.year
# Create clean DataArray with dimensions ['year', 'lat', 'lon']
datS = xr.DataArray(
data=dat_tmp.values,
dims=["year", "lat", "lon"],
coords={
"year": years.values,
"lat": dat_tmp["lat"].values,
"lon": dat_tmp["lon"].values,
},
name=dat.name if hasattr(dat, "name") else "SeasonalMean",
attrs=dat.attrs.copy(),
)
# Remove edge years if all values are missing
for yr in [datS.year.values[0], datS.year.values[-1]]:
if datS.sel(year=yr).isnull().all():
datS = datS.sel(year=datS.year != yr)
# Apply coverage mask
valid_counts = datS.count(dim="year")
total_years = datS["year"].size
min_valid_years = int(total_years * min_coverage)
sufficient_coverage = valid_counts >= min_valid_years
datS_masked = datS.where(sufficient_coverage)
dat_out = datS_masked
# Optional detrending
if dtrend:
coeffs = datS_masked.polyfit(dim="year", deg=1)
trend = xr.polyval(datS_masked["year"], coeffs.polyfit_coefficients)
dat_out = datS_masked - trend
# Update attributes
dat_out.attrs.update(datS.attrs)
dat_out.attrs["note1"] = (
f"{window}-month trailing mean ending in month {end_month}. Year corresponds to {end_month}."
)
if dtrend:
dat_out.attrs["note2"] = f"Linear trend removed after applying {int(min_coverage * 100)}% data coverage mask."
else:
dat_out.attrs["note2"] = f"Applied {int(min_coverage * 100)}% data coverage mask only (no detrending)."
return dat_out
# -- Get NDJFM mean with detrended.
dat_NDJFM = calc_seasonal_mean(dat, window=5, end_month=2, dtrend=True)
dat_MJJAS = calc_seasonal_mean(dat, window=5, end_month=9, dtrend=True)
print(dat_NDJFM)
<xarray.DataArray (year: 70, lat: 180, lon: 360)> Size: 36MB
array([[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[0.02326872, 0.02348154, 0.02298293, ..., 0.02257151,
0.02365055, 0.02348184],
[0.02483404, 0.02546713, 0.02575516, ..., 0.02538817,
0.02564338, 0.02559502],
[0.02242027, 0.0222255 , 0.02272483, ..., 0.02195308,
0.02223469, 0.02248361]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...
[0.04113557, 0.04261062, 0.04251236, ..., 0.039855 ,
0.04042611, 0.04140724],
[0.05431002, 0.05359076, 0.05437862, ..., 0.05354543,
0.05313137, 0.05407702],
[0.04739001, 0.04772313, 0.04759856, ..., 0.04785807,
0.04732536, 0.0477709 ]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[0.04385129, 0.04382134, 0.04361135, ..., 0.04428564,
0.04415513, 0.04422673],
[0.02202884, 0.02230439, 0.02168488, ..., 0.02329186,
0.02287089, 0.02251357],
[0.01074545, 0.01067164, 0.00994377, ..., 0.01071551,
0.01077672, 0.01121927]]], shape=(70, 180, 360))
Coordinates:
* year (year) int64 560B 1951 1952 1953 1954 1955 ... 2017 2018 2019 2020
* lat (lat) float32 720B -89.5 -88.5 -87.5 -86.5 ... 86.5 87.5 88.5 89.5
* lon (lon) float32 1kB 0.5 1.5 2.5 3.5 4.5 ... 356.5 357.5 358.5 359.5
Attributes:
long_name: Monthly Means of Global Sea Surface Temperature
valid_range: [-5. 40.]
units: degC
var_desc: Sea Surface Temperature
dataset: COBE-SST2 Sea Surface Temperature
statistic: Mean
parent_stat: Individual obs
level_desc: Surface
actual_range: [-3.0000002 34.392 ]
note1: 5-month trailing mean ending in month 2. Year corresponds ...
note2: Linear trend removed after applying 90% data coverage mask.
def regional_weighted_mean(data, lat1, lat2, lon1, lon2):
"""
Compute cosine-latitude-weighted regional mean over the specified lat-lon bounds.
Parameters
----------
data : xarray.DataArray
Input data with dimensions (time, lat, lon).
lat1, lat2 : float
Latitude bounds.
lon1, lon2 : float
Longitude bounds (either in -180 to 180 or 0 to 360).
Returns
-------
dat_region_mean : xarray.DataArray
Regional mean time series (time).
"""
# --- Longitude adjustment ---
target_lon_range = 'neg180_180' if lon1 < 0 or lon2 < 0 else '0_360'
if target_lon_range == 'neg180_180' and (data.lon > 180).any():
data = data.assign_coords(lon=((data.lon + 180) % 360 - 180)).sortby('lon')
elif target_lon_range == '0_360' and (data.lon < 0).any():
data = data.assign_coords(lon=(data.lon % 360)).sortby('lon')
# --- Select region ---
dat_region = data.sel(lat=slice(lat1, lat2), lon=slice(lon1, lon2))
# --- Apply cosine-latitude weighting ---
weights = np.cos(np.deg2rad(dat_region['lat']))
dat_region_mean = dat_region.weighted(weights).mean(dim=['lat', 'lon'], skipna=True)
# --- Assign name ---
dat_region_mean.name = f"regional_mean_{lat1}_{lat2}_{lon1}_{lon2}"
return dat_region_mean
# --- Calculate Niño 3.4 index from NDJFM data ---
nino34_ts = regional_weighted_mean(dat_NDJFM, -5, 5, 190, 240)
# --- Calculate standard deviation over the available years ---
nino34_std = nino34_ts.std(dim="year", skipna=True)
# --- Standardize the time series ---
nino34_ts_std = nino34_ts / nino34_std
# --- Update attributes ---
nino34_ts_std.attrs = nino34_ts.attrs.copy()
nino34_ts_std.attrs['note1'] = f"Standardized Niño 3.4 index. Standard deviation: {nino34_std.values:.3f}"
print(nino34_ts_std)
<xarray.DataArray 'regional_mean_-5_5_190_240' (year: 70)> Size: 560B
array([-0.84242396, 0.86353962, 0.15385515, 0.2855797 , -0.93414499,
-1.22521061, -0.09921426, 1.60278945, 0.58513473, 0.18864412,
-0.08125704, -0.50144525, -0.42366512, 1.01065251, -0.88651785,
1.48532399, -0.10824377, -0.47533738, 0.72837009, 0.76351155,
-1.38114218, -0.85111633, 1.68083599, -1.82370587, -0.59160717,
-1.48655636, 0.64278058, 0.7629035 , -0.07909781, 0.43672724,
-0.16109375, -0.26470221, 2.01019884, -0.70970947, -0.85609144,
-0.37170209, 1.20287726, 0.92019422, -1.73201992, -0.16373535,
0.28102254, 1.47835879, 0.08953337, 0.29129455, 0.95906713,
-1.02026181, -0.55540958, 2.35493331, -1.39454424, -1.50470693,
-0.67777684, -0.17179938, 1.15602986, 0.30964153, 0.54970597,
-0.58591287, 0.65923747, -1.67075273, -0.72687507, 1.3178921 ,
-1.60351327, -0.91385179, -0.09065725, -0.49126429, 0.54168987,
2.41709005, -0.53377439, -0.86609021, 0.7018336 , 0.4256821 ])
Coordinates:
* year (year) int64 560B 1951 1952 1953 1954 1955 ... 2017 2018 2019 2020
Attributes:
note1: Standardized Niño 3.4 index. Standard deviation: 1.038
def lead_lag_correlation_geocat(ts, ystr1, yend1, dat, ystr2, yend2, min_coverage=0.9):
"""
Compute Pearson correlation between a 1D timeseries (ts) and a gridded field (dat),
using geocat.comp.stats.pearson_r, applying a data coverage mask (e.g., 90%) and
safely skipping grid points with no variability.
Parameters
----------
ts : xr.DataArray
1D time series with dimension 'year'.
ystr1, yend1 : int
Start and end year for the timeseries.
dat : xr.DataArray
3D gridded data with dimensions ('year', 'lat', 'lon').
ystr2, yend2 : int
Start and end year for the field data.
min_coverage : float
Minimum fraction of year coverage required to compute correlation (default 0.9).
Returns
-------
corr : xr.DataArray
Correlation map with dimensions ('lat', 'lon'), masked where coverage or variability is insufficient.
"""
# Subset both datasets
ts_sel = ts.sel(year=slice(ystr1, yend1))
dat_sel = dat.sel(year=slice(ystr2, yend2))
# Check matching year sizes after subset
if ts_sel['year'].size != dat_sel['year'].size:
raise ValueError(f"Year mismatch: ts has {ts_sel['year'].size}, dat has {dat_sel['year'].size}.")
# Replace 'year' coordinate with dummy index to avoid alignment errors in geocat.comp
dummy_year = np.arange(ts_sel['year'].size)
ts_sel = ts_sel.assign_coords(year=dummy_year)
dat_sel = dat_sel.assign_coords(year=dummy_year)
# Calculate std and valid counts
dat_std = dat_sel.std(dim="year", skipna=True)
valid_counts = dat_sel.count(dim="year") # cleaner and xarray-native
min_required_years = int(dat_sel['year'].size * min_coverage)
# Create mask where std == 0 or insufficient coverage
#invalid_mask = (dat_std == 0) | (valid_counts < min_required_years)
invalid_mask = (dat_std < 1e-10) | (valid_counts < min_required_years)
# Apply mask (mask invalid areas to NaN; keep valid areas unchanged)
dat_masked = dat_sel.where(~invalid_mask)
# Calculate correlation using geocat
corr = gccomp.stats.pearson_r(ts_sel, dat_masked, dim="year", skipna=True, keep_attrs=True)
return corr
def lead_lag_correlation(ts, ystr1, yend1, dat, ystr2, yend2, min_coverage=0.9):
"""
Compute Pearson correlation between a 1D timeseries (ts) and a gridded field (dat)
using numpy's corrcoef, applying a data coverage mask (e.g., 90%).
Parameters
----------
ts : xr.DataArray
1D time series with dimension 'year'.
ystr1, yend1 : int
Start and end year for the timeseries.
dat : xr.DataArray
3D gridded data with dimensions ('year', 'lat', 'lon').
ystr2, yend2 : int
Start and end year for the field data.
min_coverage : float
Minimum fraction of year coverage required to compute correlation (default 0.9).
Returns
-------
corr_map : xr.DataArray
Correlation map with dimensions ('lat', 'lon'), masked where coverage is insufficient.
"""
# Subset both datasets for the specified years
ts_sel = ts.sel(year=slice(ystr1, yend1)).values # shape: (year,)
dat_sel = dat.sel(year=slice(ystr2, yend2)).values # shape: (year, lat, lon)
# Check matching number of years
if ts_sel.shape[0] != dat_sel.shape[0]:
raise ValueError(f"Number of years mismatch: ts has {ts_sel.shape[0]}, dat has {dat_sel.shape[0]}.")
ntime, nlat, nlon = dat_sel.shape
# Calculate valid data counts at each grid point
valid_counts = np.isfinite(dat_sel).sum(axis=0)
min_required_years = int(ntime * min_coverage)
# Mask for sufficient coverage
sufficient_mask = valid_counts >= min_required_years
# Initialize correlation map
corr_map = np.full((nlat, nlon), np.nan)
# Calculate correlation only for sufficient coverage grid points
for i in range(nlat):
for j in range(nlon):
if sufficient_mask[i, j]:
x = dat_sel[:, i, j]
mask = np.isfinite(x) & np.isfinite(ts_sel)
if mask.sum() > 2:
r = np.corrcoef(ts_sel[mask], x[mask])[0, 1]
corr_map[i, j] = r
# Convert to xarray
corr_da = xr.DataArray(
corr_map,
dims=['lat', 'lon'],
coords={'lat': dat['lat'], 'lon': dat['lon']},
name='lead_lag_correlation'
)
# Mask out insufficient data coverage explicitly
corr_da = corr_da.where(sufficient_mask)
return corr_da
corr_m12 = lead_lag_correlation_geocat(
ts=nino34_ts_std,
ystr1=1952, yend1=2020,
dat=dat_NDJFM,
ystr2=1951, yend2=2019,
min_coverage=0.9
)
corr_m6 = lead_lag_correlation_geocat(
ts=nino34_ts_std,
ystr1=1952, yend1=2020,
dat=dat_MJJAS,
ystr2=1951, yend2=2019,
min_coverage=0.9
)
corr_0 = lead_lag_correlation_geocat(
ts=nino34_ts_std,
ystr1=1951, yend1=2020,
dat=dat_NDJFM,
ystr2=1951, yend2=2020,
min_coverage=0.9
)
corr_p6 = lead_lag_correlation_geocat(
ts=nino34_ts_std,
ystr1=1951, yend1=2020,
dat=dat_MJJAS,
ystr2=1951, yend2=2020,
min_coverage=0.9
)
/Users/a02235045/miniconda3/envs/vbook/lib/python3.13/site-packages/xskillscore/core/np_deterministic.py:314: RuntimeWarning: invalid value encountered in divide
r = r_num / r_den
/Users/a02235045/miniconda3/envs/vbook/lib/python3.13/site-packages/xskillscore/core/np_deterministic.py:314: RuntimeWarning: invalid value encountered in divide
r = r_num / r_den
/Users/a02235045/miniconda3/envs/vbook/lib/python3.13/site-packages/xskillscore/core/np_deterministic.py:314: RuntimeWarning: invalid value encountered in divide
r = r_num / r_den
/Users/a02235045/miniconda3/envs/vbook/lib/python3.13/site-packages/xskillscore/core/np_deterministic.py:314: RuntimeWarning: invalid value encountered in divide
r = r_num / r_den
def plot_global_panel_OP2(ax, std_data, std_levels, titleM, titleL, color_list,
clm_data=None, clm_levels=None):
"""
Plot global map of monthly standard deviation with optional climatology contours.
Parameters
----------
ax : cartopy.mpl.geoaxes.GeoAxesSubplot
Axes to plot on.
std_data : xarray.DataArray (month, lat, lon)
Standard deviation data to plot with filled contours.
std_levels : list or array-like
Contour levels for the std_data.
title : str
Title above the plot panel.
color_list : list
Colors for the colormap.
clm_data : xarray.DataArray (month, lat, lon), optional
Climatology data for contour overlay.
clm_levels : list or array-like, optional
Contour levels for climatology.
"""
# --- Add cyclic longitudes ---
std_cyclic = gvutil.xr_add_cyclic_longitudes(std_data, "lon")
# --- Plot standard deviation ---
im = ax.contourf(
std_cyclic["lon"],
std_cyclic["lat"],
std_cyclic,
levels=std_levels,
cmap=color_list,
extend="both",
transform=ccrs.PlateCarree()
)
# --- Plot climatology contours (optional) ---
if clm_data is not None:
clm_cyclic = gvutil.xr_add_cyclic_longitudes(clm_data, "lon")
levels = clm_levels if clm_levels is not None else 10
csW = ax.contour(
clm_cyclic["lon"], clm_cyclic["lat"], clm_cyclic,
levels=levels, colors="white", linewidths=3,
transform=ccrs.PlateCarree()
)
csB = ax.contour(
clm_cyclic["lon"], clm_cyclic["lat"], clm_cyclic,
levels=levels, colors="black", linewidths=1,
transform=ccrs.PlateCarree()
)
ax.clabel(csW, inline=1, fontsize=8, fmt="%.0f")
ax.clabel(csB, inline=1, fontsize=8, fmt="%.0f")
# --- Map features ---
ax.add_feature(cfeature.LAND, facecolor='lightgray', edgecolor='none', zorder=0)
ax.coastlines(linewidth=0.5, alpha=0.6)
ax.set_extent([-180, 180, -90, 90], crs=ccrs.PlateCarree())
# --- Option 2: Manual setting ---
# --- Set major ticks explicitly ---
ax.set_xticks(np.arange(-180, 181, 60), crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(-90, 91, 30), crs=ccrs.PlateCarree())
# Custom tick labels
ax.set_yticklabels(["90°S", "60°S", "30°S", "EQ", "30°N", "60°N", "90°N"])
ax.set_xticklabels(["180°", "120°W", "60°W", "0°", "60°E", "120°E", ""])
# --- Set minor tick locators ---
ax.xaxis.set_minor_locator(mticker.MultipleLocator(15))
ax.yaxis.set_minor_locator(mticker.MultipleLocator(10))
# --- Add tick marks ---
gvutil.add_major_minor_ticks(
ax,
labelsize=10,
x_minor_per_major=4, # 15° spacing for minor if major is 60°
y_minor_per_major=3 # 10° spacing for minor if major is 30°
)
# --- Customize ticks to show outward ---
ax.tick_params(
axis="both",
which="both",
direction="out",
length=6,
width=1,
top=True,
bottom=True,
left=True,
right=True
)
ax.tick_params(
axis="both",
which="minor",
length=4,
width=0.75
)
gvutil.set_titles_and_labels(ax, maintitle=titleM, maintitlefontsize=16, lefttitle=titleL, lefttitlefontsize=14)
return im
# --- Global settings ---
proj = ccrs.PlateCarree(central_longitude=200)
fig = plt.figure(figsize=(12, 11))
gs = gridspec.GridSpec(nrows=3, ncols=8, height_ratios=[1.0, 1.0, 1.0], hspace=0.5, wspace=1.0)
ax0 = fig.add_subplot(gs[0, 1:7])
ax1 = fig.add_subplot(gs[1, 0:4], projection=proj)
ax2 = fig.add_subplot(gs[1, 4:8], projection=proj)
ax3 = fig.add_subplot(gs[2, 0:4], projection=proj)
ax4 = fig.add_subplot(gs[2, 4:8], projection=proj)
#fig.suptitle("SST Anomalies during ENSO events", fontsize=16, fontweight="bold")
colormap = cmaps.BlueYellowRed.colors # Use the AMWG BlueYellowRed colormap
color_list = ListedColormap(colormap)
color_list.set_under(colormap[0])
color_list.set_over(colormap[-1])
clevs_corr = np.arange(-0.9, 1.0, 0.1)
# === Top panel: Niño 3.4 index ===
def setup_axis(ax, ystr, yend, ymin, ymax, y_major, y_minor):
# Make grid go below the bars
ax.set_axisbelow(True)
# X-Axis
#ax.xaxis.set_major_locator(MultipleLocator(10))
ax.xaxis.set_minor_locator(MultipleLocator(1))
ax.set_xlim(ystr,yend)
# Y-Axis
ax.set_ylim(ymin, ymax)
ax.yaxis.set_minor_locator(MultipleLocator(y_minor))
# Customized grid setting
ax.tick_params(axis="both", which="major", length=7, top=True, right=True)
ax.tick_params(axis="both", which="minor", length=4, top=True, right=True)
ax.grid(visible=True, which="major", linestyle="-", linewidth=0.7, alpha=0.7)
ax.grid(visible=True, which="minor", linestyle="--", linewidth=0.4, alpha=0.5)
positive_color = "pink"
negative_color = "lightblue"
positive_color_edge = "red"
negative_color_edge = "blue"
bar_colors = [positive_color if val >= 0 else negative_color for val in nino34_ts_std]
edge_colors = [positive_color_edge if val >= 0 else negative_color_edge for val in nino34_ts_std]
ax0.bar(nino34_ts_std.year, nino34_ts_std, color=bar_colors, edgecolor=edge_colors,
linewidth=1, label="Nino 3.4 index")
setup_axis(ax0, ystr, yend+1, -3., 3., 1, 0.2)
gvutil.set_titles_and_labels(ax0,
maintitle="SST Anomalies during ENSO events",
lefttitle="(a) Nino 3.4 Index in NDJFM",
righttitle="",
ylabel="Normalized",
xlabel="Year",
maintitlefontsize=14, # Adjust main title font size
lefttitlefontsize=12, # Set left title font size
righttitlefontsize=12, # Set left title font size
labelfontsize=10 # Set y-label font size
)
ax0.axhline(0, color="gray", linestyle="-")
# === Map panels ===
im1 = plot_global_panel_OP2(ax1, std_data=corr_m12, std_levels=clevs_corr,
titleM="", titleL="(a) SST anomalies at -12 month",
color_list=color_list)
im2 = plot_global_panel_OP2(ax2, std_data=corr_m6, std_levels=clevs_corr,
titleM="", titleL="(b) SST anomalies at -6 month",
color_list=color_list)
im3 = plot_global_panel_OP2(ax3, std_data=corr_0, std_levels=clevs_corr,
titleM="", titleL="(c) SST anomalies at 0 month",
color_list=color_list)
im4 = plot_global_panel_OP2(ax4, std_data=corr_p6, std_levels=clevs_corr,
titleM="", titleL="(d) SST anomalies at +6 month",
color_list=color_list)
# --- Add a single shared colorbar for the maps ---
cbar_ax = fig.add_axes([0.25, 0.05, 0.5, 0.02])
cbar = plt.colorbar(im1, cax=cbar_ax, orientation="horizontal",
spacing="uniform", extend='both', extendfrac=0.07,
drawedges=True)
cbar.set_label("Correlation Coefficient", fontsize=12)
cbar.ax.tick_params(length=0, labelsize=10)
cbar.outline.set_edgecolor("black")
cbar.outline.set_linewidth(1.0)
# --- Finalize ---
plt.savefig(fnFIG, dpi=300, bbox_inches="tight")
plt.show()